Lasers Identify Key Molecular Structure in Tumours
Interview with
Ben - Also in the news this week, a laser technique has exposed the previously unknown molecular shape of Epidermal Growth Factor Receptors or EGFRs which are known to be involved in the development of cancer. These are found on the surface of the vast majority of human cells as Dr. Marisa Martin-Fernandez, a scientist based at the STFC's Central Laser Facility in Harwell explained...
Marisa - The role of this receptor is to bind molecules which are in the bloodstream and produce signals which are transduced to the inside of each cell and to give instructions to the cell machinery to grow, divide, die, differentiate. So it's kind of the core of how to promote cell behaviour. These are the orchestrators, these type of receptors orchestrate what cells are going to do within a multicellular body and there is a lot that is known about what happens in this signalling process that leads to cellular growth and when it goes wrong and leads to the growth of tumours. In fact, most human tumours have a fault in the signalling of this receptor.
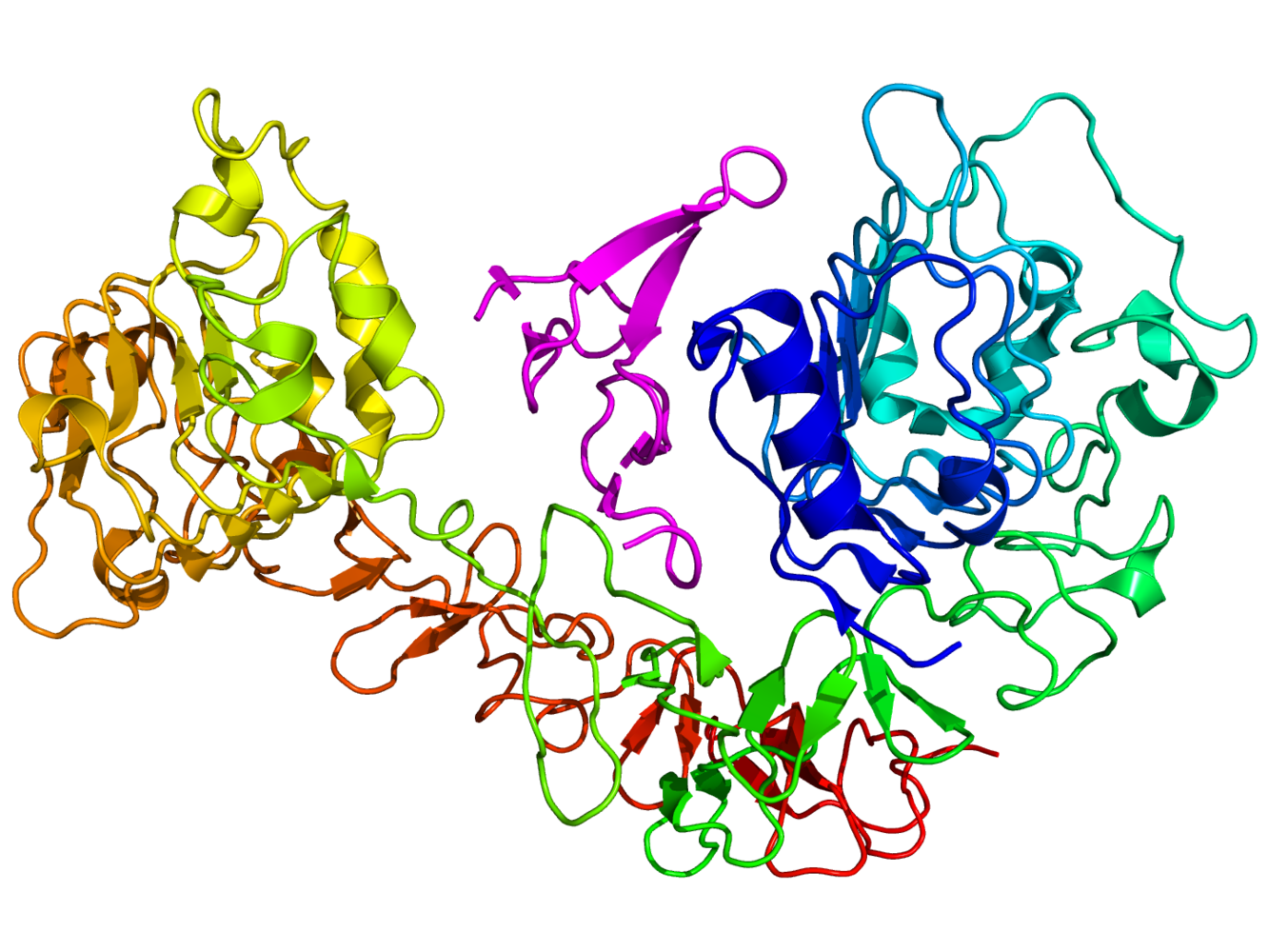 Ben - Current drugs that target EGFRs do so non-discriminately, blocking activation to halt cell growth. However, in doing so, they may block other essential cell maintenance and this could lead to the body adopting other chemical pathways to achieve the same goal. This side steps the cancer treatment and allows the tumour to grow once more. To find out more about the precise interactions between EGFRs and signalling molecules, we need a good understanding of their structure. To do this, Marisa's team using novel laser technique called fluorescence resonance energy transfer...
Ben - Current drugs that target EGFRs do so non-discriminately, blocking activation to halt cell growth. However, in doing so, they may block other essential cell maintenance and this could lead to the body adopting other chemical pathways to achieve the same goal. This side steps the cancer treatment and allows the tumour to grow once more. To find out more about the precise interactions between EGFRs and signalling molecules, we need a good understanding of their structure. To do this, Marisa's team using novel laser technique called fluorescence resonance energy transfer...
Marisa - So what we do is we put a label in a position in the receptor that we can control, which is a molecule that emits fluorescence, and we put another label on the cell surface, and then we excite one of the molecules in the receptor. Depending on how close it is to the surface, the characteristics of the fluorescence emission are different and from that information, we can actually get nanometer distances.
Ben - Knowing the distances between the receptor and the other parts of the cell can help to simulate exactly what structure the receptor will take in situ. This job is taken on by experts such as Dr. Martyn Winn of the STFC's Computational Science and Engineering Department at the Daresbury laboratory...
Martyn - Our starting points are atomic structures that you get from crystallography. These are experiments that would take place on Diamond Light Source, for example, and they give you very detailed atomic structures of the proteins that are involved. The drawback of that is they're static. They don't move around like they do in real life. Also, that means that the proteins have been taken out of the cell, taken out of their natural context and put into a crystal. So you have very detailed information but it's not necessarily relevant to what's happening in the cell. So what we would do in a simulation is to take that detailed structure, set up a simulation which mimics the cell environment and then see what happens when it's put into the cellular context.
Ben - When Marisa's laser data was combined in a simulation with the known atomic structure of EGFRs, the resulting shape was new, unexpected, but surprisingly similar to a structure found in Drosophila, the fruit fly.
Martyn - So what you often see from crystallography is very symmetric structures. The crystal environment is a very symmetric environment and everything is nice and well ordered. What we saw when we did our simulation was that the molecule actually becomes asymmetric. When we first did this, this was something that had not been seen before and slightly heretic. People love symmetry. Symmetry is beautiful, but our simulations were showing that the molecule became asymmetric in order to explain Marisa's experiments.
Marisa - And then the amazing thing is that when we look at the molecular shape, it looked nearly identical to the molecular shape of the Drosophila receptor. And then when we identified the shape, it was virtually identical. It was so similar, it was actually uncanny. We were amazed, when we looked at the structure that we identified, and it looked like the Drosophila receptor, we knew we were onto something.
Ben - So, knowing this particular structure could help lead to better therapeutics and the technique itself could help move forward into personalised medicine.
Marisa - Given this new information, we could actually consider new antibody therapeutics that could block one of the other version of the receptor and allow the other signals to be transduced. So, in a way, they will be less invasive therapeutics that might actually reduce side effects, and make sure that, for example, the body is not deprived of a fundamental function that this receptor covers.
Martyn - And I think we're at the level of very basic science and it's a long road towards drug development, that's true, but I think one of the things that they want to do is to develop treatments that are specific to patients. So you can analyse the DNA from particular patients to see if they have particular mutations. It might be that you can also analyse the tumours of patients to see what the confirmation of the receptors are and then that might shed light on whether a particular antibody treatment is likely to work or not. That their interest, actually in implying this to patient-specific treatments.
Ben - That was Martyn Winn from the Daresbury Lab and Marisa Martin-Fernandez from the Central Laser Facility in Harwell. You can find that paper in the journal Molecular and Cellular Biology.





Comments
Add a comment